Frequently Asked Questions
1. What species does BioGPS support?
Currently, BioGPS supports eight species: Human (Homo sapiens), Mouse (Mus musculus), Rat (Rattus norvegicus), Fruitfly (Drosophila melanogaster), Nematode (Caenorhabditis elegans), Zebrafish (Danio rerio), Thale-cress (Arabidopsis thaliana), Frog (Xenopus tropicalis), and Pig (Sus scrofa). BioGPS presents data in an ortholog-centric format, which allows users to display mouse plugins next to human ones. Our data for defining orthologs comes from NCBI's HomoloGene database.
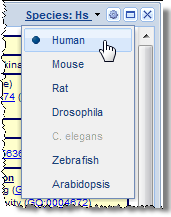
For plugins that contain data for multiple species, users can switch between allowed species using the species selector in the upper-right corner of the plugin window, as shown at right. Species which are grayed-out mean either that the species does not have an ortholog for that gene, or that the selected plugin does not provide data for that species.
2. How do I cite BioGPS in my paper?
Manuscripts describing BioGPS have been published in the database issues of Nucleic Acid Research in 2016 and 2013, as well as in Genome Biology. Note that when referencing any of the expression data sets in BioGPS, you should also include a citation to the relevant paper describing those data. Please see the Publications section on our Downloads page.
3. How do I deep link to BioGPS?
Deep links (or permalinks) are most easily generated based on Entrez Gene ID. For example, for the gene CDK2 (Entrez Gene ID = 1017):
Queries based on identifiers can also be constructed using URLs like this:
- http://biogps.org/?query=CDK2 (searches by symbols, identifiers, and aliases only)
- http://biogps.org/?query=CDK2&qtype=keyword (searches all text fields)
Results from a specific dataset can be constructed using URLs like this:
- http://biogps.org/#goto=genereport&id=12566&show_dataset=E-GEOD-45045 (provides results for a specific gene from a specific data set
If additional linking mechanisms are required, please contact us.
4. What features are available for users who have created an account?
| Anonymous | Account | |
|---|---|---|
| Search | ||
| View default gene report layouts | ||
| Customize layouts | ||
| Register new plugins | ||
| Save gene lists |
5. What kind of gene identifiers are supported for a query?
In addition to official symbols and aliases, BioGPS supports most common public gene identifiers, including Entrez Gene, Ensembl, Uniprot, Refseq, UniGene, Gene Ontology, PDB, and Interpro. In addition, users can search by Affymetrix probe set identifiers from many common gene expression chip types (U133, MOE430, U95, RGU34, GNF1H and GNF1M).
6. Help! There are too many scrollbars!
Although not technically a question, this is a frequently heard frustration. Undoubtedly BioGPS works better on higher-resolution monitors, but we also have some pretty effective strategies for limiting scrollbar overload for all users.
- First, use fewer plugins per layout. In fact, we often create layouts with just a single plugin.
- Second, use the maximize plugin button in the upper right of the plugin window, which will show one plugin in a large size while freezing the main browser scrollbar.
- Third, use the "Open in browser" option under the "more options" plugin menu. This option will open the plugin in a new browser tab or window.
- Fourth, use the "Open all in browser" feature under the layout options menu. Selecting this option will load all plugins, each in its own browser tab or window.

Gene Expression Charts
7. What are the units of expression?
Expression values from Affymetrix chips relate to fluorescence intensity. Since there are multiple probes for each transcript on the microarray, these intensity values are summarized using various data processing algorithms. In our case, we mostly use gcrma.
8. Can I compare values across tissues? across probe sets?
Since each probe and probe set has different background characteristics (depending of cross hybridization, secondary structure, etc.), comparing expression values across different probesets in a single sample is generally not recommended. Microarrays are primarily suited for comparing single probesets across multiple samples.
9. Help, I can't see the gene expression chart for my gene, or the text is too small.
For known issues and solutions regarding the gene expression charts, please check out this thread:
Please see here for known bugs and work arounds with regards to viewing the Gene Expression Chart:
in the biogps google group
If you are still having problems, please post/send a screenshot and include the name of your Browser
(with version #) and OS in the your help request so we can replicate the issue and solve the problem.
Datasets
10. Where did the datasets come from and how was the data obtained?
There are multiple datasets used by BioGPS. For more detailed information on how the data from each dataset was generated, you can view/download the associated peer-reviewed publication from the Publications and References section on the following page: http://biogps.org/downloads/.
11. Where can I purchase or learn more about the exact cell/tissue/sample seen in your datasets?
Good question. Your best bet would be to find the publication associated with the dataset where your exact cell/tissue/sample is found, and inquire with the author of that publication.
12. What is the difference between an "averaged" and "unaveraged" dataset files? Are the values from the "averaged" dataset files normalized?
The averaged values are normalized, and different datasets may have been normalized differently. See question #7 for more info.
13. I am only interested in gene expression from a specific cell/tissue/organ/system. How can I get all the gene expression data for only my cell/tissue/organ/system of interest?
Since there is no exact criteria for specifying cell/tissue/organ/system, there is no official way to do this through bioGPS at this time. Additionally, you should be careful when comparing expression data across different data sets since different probes may have been used, or different methodologies applied. You can, however, search for datasets which better suit your purpose by searching for the appropriate dataset tag.
14. BioGPS is not working. What should I do?
In order for us to help you, we need the right information from you. Please follow the instructions in the BioGPS self-help guide before contacting us as this will allow us to better help you.
